Crystallography of macromolecules
X-ray crystallography allows a “look” at the detailed (atom-by-atom) structure of molecules in 3 dimensions. For this reason, it has an immense impact on chemistry, material science and biology. It has generated a whole new field in the latter, “Structural Biology”. Knowledge of the structure of biological macromolecules can explain their function (or malfunction), reveal mechanisms and pathways in the cells and eventually allow us to understand life and use this knowledge to our benefit, e.g. to improve the design of drugs.
- Past projects include elucidation of the structures of Glucogen Phosphorylase b with γ-CD,1 several 2[4Fe4S] ferredoxins from pathogenic bacteria,2-3 certain domains of the muscle proteins titin4 and myomesin.5
- Endoplasmic Reticulum Aminopeptidases: ER aminopeptidases ERAP1 and ERAP2, as well as the insulin-regulated aminopeptidase IRAP, cooperate to trim a vast variety of antigenic peptide precursors to generate mature epitopes, thus regulating the immune response. In collaboration with the group of E. Stratikos, we have determined the structure of ERAP2 by X-ray crystallography.6 The structure helps explain how two homologous aminopeptidases cooperate to process a large variety of sequences, a key property to their biological role.
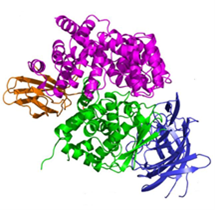
Common coding single nucleotide polymorphisms (SNPs) in ERAP1 and ERAP2 have been linked with predisposition to human diseases ranging from viral infections to autoimmunity and cancer. A common ERAP2 SNP coding for amino acid variation N392K, leads to alterations in the activity and specificity of the enzyme. X-ray crystallographic analysis of the 392K allele provides insight to the association of this ERAP2 polymorphism with disease and supports the idea that polymorphic variation in antigen processing enzymes constitutes a component of immune response variability in humans.7
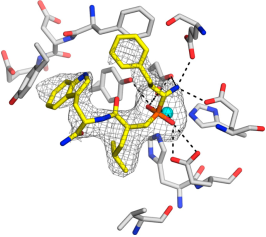
The structures of several ERAP2 and IRAP complexes with rationally designed inhibitors (drug candidates) have also been determined. Thus insight is gained on the structural requirements for an inhibitor to offer better binding affinity to the protein, which can provide lead drug candidates.8-13
- DsbD: a bacterial thiol-disulfide oxidoreductase: DsbD, one of the five proteins of the bacterial Disulfide bond (Dsb) formation system, plays an important role in oxidative protein folding allowing the correct formation of disulfide bonds in proteins. Located in the inner membrane of Gram-negative bacteria, it is responsible for transporting reducing power from the cytoplasm to the oxidising periplasm of the cell. DsbD is involved in bacterial pathogenesis as the expression and stability of most virulent factors are dependent on its presence. Four structures of the soluble domains of the transmembrane protein DsbD have been solved: the N-terminal domain in the reduced form14, the C-terminal domains of two point-mutant variants15, and a deletion mutant of the oxidized N-terminal domain,16 in collaboration with the Department of Biochemistry, University of Oxford (Prof. S. Ferguson, Prof. C. Redfield and Dr. D. Mavridou). This work contributed significantly to the elucidation of the interactions of the soluble domains of this unique oxidoreductase, but also in the general understanding of the factors controlling the reactivity of the ubiquitous thioredoxin fold.
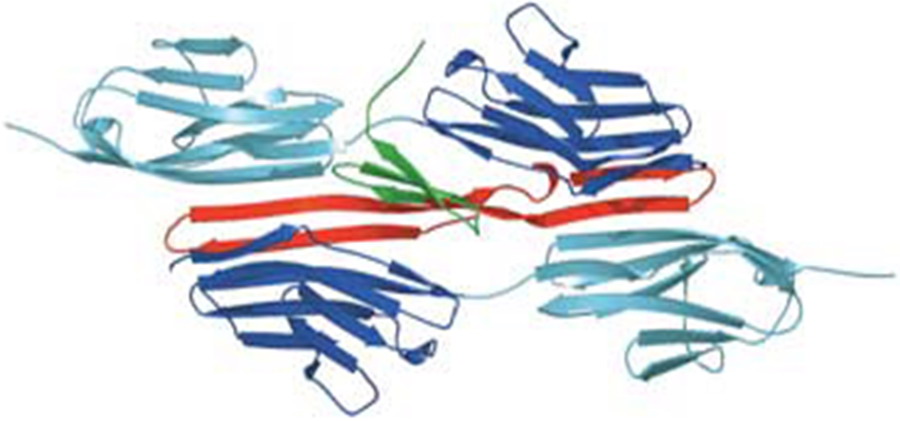
- Anti-CCR5 antibody RoAb13 in complex with epitope: C-C chemokine receptor 5 (CCR5) is a major co-receptor molecule used by HIV-1 to enter cells, causing AIDS. This led to the hypothesis that stimulating an antibody response would block HIV with minimal toxicity. X-ray crystallographic studies of the anti-CCR5 antibody RoAb13 alone and in complex with CCR5 epitope sequence peptides were undertaken, in a collaboration including groups at University College London, Imperial College London and the University of Manchester.17,18 The peptides were observed to bind at a site within a hypervariable CDR3 binding region. These structures may prove useful in the design of a new biomimetic drug to stimulate an immune response to CCR5 in order to block HIV infection.
Related references
- The binding of beta and gamma-cyclodextrins to glycogen phosphorylase b: kinetic and crystallographic studies. N. Pinotsis, D. D. Leonidas, E. D. Chrysina, N. G. Oikonomakos, I. M.Mavridis Protein Sci. 2003, 12, 1914-1924.
- The structure of the 2[4Fe-4S] ferredoxin from Pseudomonas aeruginosa at 1.32 Å resolution. Comparison with other high resolution structures of ferredoxins and contributing structural features to reduction potential values. P. Giastas, N. Pinotsis, G. Efthymiou, M. Wilmanns, P. Kyritsis, J-M. Moulis, I. M. Mavridis J. Biolog. Inorg. Chem. 2006,11, 445-458.
- Insight into the protein and solvent contributions to the reduction potentials of [4Fe-4S]2+/+ clusters: Crystal structures of the Allochromatium vinosum ferredoxin variants C57A and V13G and the homologous Escherichia coli ferredoxin. E. Saridakis, P. Giastas, G. Efthymiou, V. Thoma, J-M. Moulis, P. Kyritsis, I. M. Mavridis J. Biolog. Inorg. Chem. 2009, 14, 783-799.
- Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. P. Zou, N. Pinotsis, S. Lange, Y-H. Song, A. Popov, I. Mavridis, O. M. Mayans, M. Gautel, M. Wilmanns, Nature 2006, 439, 225-228.
- Superhelical architecture of the myosin filament-linking protein myomesin with unusual elastic properties. N. Pinotsis, S. D. Chatziefthimiou, F. Berkemeier, F. Beuron, I. M. Mavridis, P. V. Konarev, D. I. Svergun, E. Morris, M. Rief, M. Wilmanns, PLoS Biol. 2012, Feb;10(2):e1001261.
- Crystal Structure of Human ER Aminopeptidase 2 Reveals Atomic Basis for Distinct Roles in Antigen Processing. R. Birtley, E. Saridakis, E. Stratikos, I. M. Mavridis Biochemistry, 2012, 51, 286-295.
- A Common Single Nucleotide Polymorphism in Endoplasmic Reticulum Aminopeptidase 2 Induces a Specificity Switch that Leads to Altered Antigen Processing. D. Evnouchidou, J. Birtley, S. Seregin, A. Papakyriakou, E. Zervoudi, M. Samiotaki, G. Panayotou, P. Giastas, O. Petrakis, D. Georgiadis, A. Amalfitano, E. Saridakis, I. M. Mavridis, E. Stratikos, J. Immunol. 2012, 189, 2383-2392.
- A rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses. E. Zervoudi, E. Saridakis, J.R. Birtley, S. Seregin , E. Reeves, P. Kokkala, Y.A. Aldhamen, A. Amalfitano, I.M. Mavridis, E. James, D. Georgiadis and E. Stratikos, Proc. Natl. Acad. Sci. USA 2013, 110, 19890-19895.
- Structural basis for antigenic peptide recognition and processing by ER aminopeptidase 2. A. Mpakali, P. Giastas, N. Mathioudakis, I.M. Mavridis, E. Saridakis, E. Stratikos J. Biol. Chem. 2015, 290, 26021-26032.
- Crystal structure of Insulin-Regulated Aminopeptidase with bound substrate analogue provides insight on antigenic epitope precursor recognition and processing. A. Mpakali, E. Saridakis, K. Harlos, Y. Zhao, A. Papakyriakou, P. Kokkala, D. Georgiadis, E. Stratikos, J. Immunol. 2015, 195, 2842-2851.
- Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency. A. Mpakali, P. Giastas, R. Deprez-Poulain, A. Papakyriakou, D. Koumantou, R. Gealageas, S. Tsoukalidou, D. Vourloumis, I. M Mavridis, E. Stratikos, E. Saridakis, ACS Med. Chem. Lett. 2017, 8 (3), 333-337.
- Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. A. Mpakali, E. Saridakis, K. Harlos, Y. Zhao, P. Kokkala, D. Georgiadis, P. Giastas, A. Papakyriakou, E. Stratikos, J. Med. Chem. 2017, 60, 2963-2972.
- Structural basis of inhibition of insulin-regulated aminopeptidase by a macrocyclic peptidic inhibitor. A. Mpakali, E. Saridakis, P. Giastas, Z. Maben, L.J. Stern, M. Larhed, M. Hallberg and E. Stratikos. ACS Med. Chem. Lett.2020, 11, 1429-1434.
- Oxidation-state-dependent protein-protein interactions in disulfide cascades. D. A. I. Mavridou, E. Saridakis, P. Kritsiligkou, A.D. Goddard, J.M. Stevens, S.J. Ferguson and C. Redfield. J. Biol. Chem. 2011, 286, 24943-24956.
- An extended active-site motif controls the reactivity of the thioredoxin fold. D. A. I. Mavridou, E. Saridakis, P. Kritsiligkou, E.C. Mozley, S.J. Ferguson and C. Redfield, J. Biol. Chem. 2014, 289, 8681-8696.
- Local frustration determines loop opening during the catalytic cycle of an oxidoreductase. L. S. Stelzl, D. A. I. Mavridou, E. Saridakis, D. Gonzalez, A. J. Baldwin, S. J. Ferguson, M. S. P. Sansom and C. Redfield, eLife 2020, 9:e54661.
- A Linear Epitope in the N-Terminal Domain of CCR5 and Its Interaction with Antibody. B. Chain, J. Arnold, S. Akthar, M. Brandt, D. Davis, M. Noursadeghi, T. Lapp, C. Ji, S. Sankuratri, Y. Zhang, L. Govada, E. Saridakis and N. Chayen, PLoS One 2015, 10 (6), e0128381.
- X-ray crystallographic studies of RoAb13 bound to PIYDIN, a part of the N-terminal domain of C-C chemokine receptor 5. L. Govada, E. Saridakis, S. C. Kassen, A. Bin-Ramzi, R. M. Morgan, B. Chain, J. R. Helliwell and N. E. Chayen, IUCrJ 2021, 8, 678-683.